|
The project described here has also been published in the journal Mechanisms of Development, 100 (2001) pp. 59-63. The pages here contain extra information not available in the paper.
| Abstract |
3-dimensional computer reconstructions of gene
expression data will become a valuable tool in biomedical research in the
near future. However, at present the process of converting in-situ expression
data into 3D models is a highly specialised and time-consuming procedure.
Here we present a method which allows rapid reconstruction of wholemount
in-situ data from mouse embryos. Mid-gestation embryos were stained with
the alkaline phosphotase substrate Fast Red, which can be detected using
confocal laser scanning microscopy (CLSM), and cut into 70 micron sections.
Each section was then scanned individually and digitally reconstructed.
Using this method it took one working day to section, digitise and reconstruct
the full expression pattern of Shh in an E9.5 embryo. Aditionally we demonstrate
that this technique allows gene expression to be studied at the single
cell level in intact tissue.
| Movies |
- Reconstruction of the full expression pattern of Shh in a 9.5dpc embryo - mpg.
- Frontal slices through the Shh reconstruction - mpg.
- Optical slices through a single thick section of the floorplate and notochord stained for Shh - mpg.
| Recreating accurate shapes in 3D |
A consistent issue affecting all serial-section reconstruction projects, is how to ensure that the reconstructed 3D shapes of organs accurately represent the real shapes within the embryo. There are at least two components to this problem. The first of these is that the sections may deform during the cutting or mounting process. This is particularly true for parafin wax sections, which tend to crumple during cutting, and then stretch out again while being floated on warm water for mounting. The second problem is how to align the sections once they have all been digitised.The most direct solution to the first problem, is to use a cutting technique which creates more robust sections. Streicher et al. embed their embryos in resin instead of parafin wax, as this can produce sections which are still thin (5-7 microns), but less fragile and much less deformable. The alternative, presented here, is to cut thick sections. Retaining a larger bulk of intact tissue within each section makes the resultant block resistant to being stretched. The improved alignment of internal structures, compared to wax sections, could clearly be seen by the fact that "warping" (described next) was unnecessary to create the reconstruction.
The simplest method for aligning serial sections depends on the fact that adjacent sections will always appear anatomically similar. A series of landmark points (tie-points) are selected by hand on both of the sections, which indicate the same anatomical feature on each, and the transformation which best matches the two sets of landmarks (a combination of rotation and translation) is calculated by a computer program. In some reconstruction projects (such as the Mouse Atlas Project), this is augmented by a more complex transformation procedure, known as warping, in which each section is treated like an elastic sheet, and a degree of local deformations is allowed. Tie-points are again used to define equivalent positions on adjacent slides, and the resultant transformation compensates for the deformations caused during the cutting and mounting stage.
These fitting procedures will produce shapes which are smooth and coherent, but not necessarily the same as the original ones. A degree of 'straightening' is inevitable, in which structures whose boundaries are diagonal to the vertical axis, become more parallel to it (Figure1). This is because each thin section is abstracted to a 2D object with no information about the angle at which structures pass through it. Matching tie-points as closely as possible ignores the fact that some of these structures should not in fact be lined-up, but should be slightly displaced. The resultant distortion will be reduced by the presence of neighbouring structures which traverse the sections in other orientations, however it can never be confidently eliminated without extra information.
There are a number of different ways to provide this extra information. One method is to embed embryos is resin and then drill vertical holes through the block before sectioning Streicher et al., 2000). These act as high-reliability fudicial markers, and are used by specially-written computer software to automatically align the sections. This technique also depends on the fact that the resin-embedded embryos do not deform much during the cutting stage. Another option is to take a digital image of the cut end of the block before each section is cut, thereby capturing a perfect representation of the original shapes within the block.
Thick sections have a theoretical advantage over all thin-section techniques in that they contain edge-gradient information, rather than only edge-position information. This extra level of information derives from the fact that the optical sections within them are perfectly aligned. It means that the correct shape can be retrieved without resorting to one of the external sources of alignment information mentioned above. The easiest way to take advantage of this information is simply to align the image at the bottom of one section with the image at the top of the adjacent section (Figure 1). Using this approach we were able to reconstruct the rounded shape of the ZPA region of an E10.5 limb bud (Figure 2). This would have been impossible from thin sections (again, unless one of the extra sources of alignment data mentioned above was also used), because the optical sections all appear very similar, and there are no internal organs which could provide anatomical landmarks for tie-points. The only distinguishable features were positions on the ectoderm-mesenchyme boundary, and while they were enough to align the thick sections, using them to align thin sections would have caused the rounded shape to be straightened out, in a manner similar to the thin-sectioned tube in Figure 1.
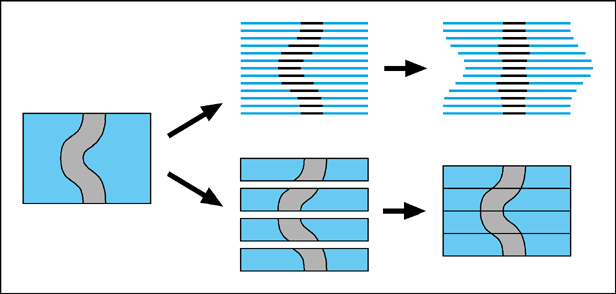
The bottom two diagrams illustrate how 3D information is preserved in thick sections.
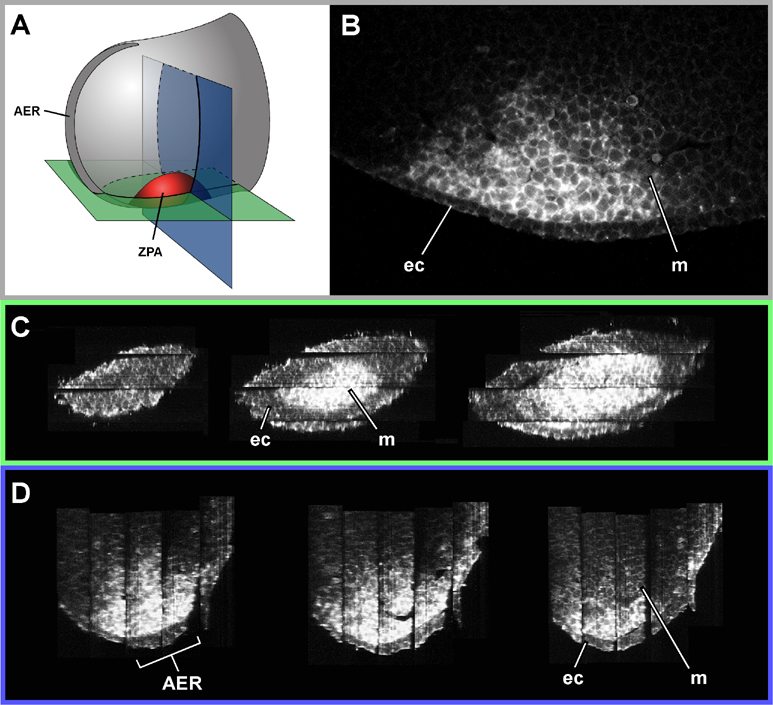
Figure 2. High resolution reconstruction of the ZPA (Zone of Polarising Activity) of a 10.5dpc embryo stained for Shh. A) Diagram showing the orientation of virtual sections (shown in C and D)
B) One of the original optical sections (perpemdicular to C and D)
C) Virtual sections which start at the posterior end of the limb bud, and slice progressively anteriorly, cutting first through the ectoderm, and then through the ZPA
D) Virtual sections which start distally (where it cuts through the AER - Apical Ectodermal Ridge) and ends more proximally
ec - ectoderm, m - mesenchyme